-
Schuster, Stefan, Univ.-Prof. Dr Professorship of Bioinformatics
Room 3403
Ernst-Abbe-Platz 1-2
07743 Jena
-
Dimitriew, Wassili Professorship of Bioinformatics
Room 3427
Ernst-Abbe-Platz 1-2
07743 Jena
-
Project description
Organisms and their traits are shaped by evolution leading to an optimized metabolism and cell behaviour. In this project, we use dynamic optimization, a widely used tool in engineering, to understand the optimality principles in biological systems with respect to time, which is often neglected by other approaches. Specifically, we want to understand the principles behind the regulation of cell metabolism and the interaction of pathogenic microorganisms with the immune system. Due to the embedding of the project in the Transregio "FungiNet", a close cooperation with experimentalists ensures the validation of the gathered hypotheses.
-
Literature
- W. Dimitriew, S. Schuster
Dynamic optimization elucidates higher-level pathogenicity strategies of Pseudomonas aeruginosa
µLife, 2025, accepted - J. Ewald, F. Rivieccio, L. Radosa, S. Schuster, A.A. Brakhage, C. Kaleta
Dynamic optimization reveals alveolar epithelial cells as key mediators of host defense in invasive aspergillosis
PLOS Comp. Biol. 17, 2021, e1009645 - Ewald, M. Bartl, C. Kaleta
Deciphering the regulation of metabolism with dynamic optimization: an overview of recent advances
Biochemical Society Transactions 45 (4), 2017, 1-9 - Ewald, M. Bartl, T. Dandekar, C. Kaleta
Optimality principles reveal a complex interplay of intermediate toxicity and kinetic efficiency in the regulation of prokaryotic metabolism
PLOS Computational Biology 13, 2017, e100537 - F. Wessely, M. Bartl, R. Guthke, P. Li, S. Schuster, C. Kaleta
Optimal regulatory strategies for metabolic pathways in Escherichia coli depending on protein costs.
Molecular Systems Biology 7, 2011, 515
- W. Dimitriew, S. Schuster
-
Funding
DFG Collaborative Research Center / Transregio 124 "FungiNet"
-
Project description
A cornerstone of microbial pathogenicity is the flexibility and robustness of their metabolism, which allows pathogens to survive and grow within the host. In this project mathematical and computational approaches are combined to find possible weak points in the metabolism of pathogens. Particularly, toxic intermediates in fungal specific pathways are investigated to explore new antifungal intervention strategies against pathogenic fungi like Candida albicans.
-
Literature
- J. Ewald, P. Sieber, R. Garde, S.N. Lang, S. Schuster, B. Ibrahim
Trends in mathematical modeling of host-pathogen interactions
Cell. Mol. Life Sci. Volume 77, Issue 3, 2020, pp 467–480 - Ewald, M. Bartl, T. Dandekar, C. Kaleta
Optimality principles reveal a complex interplay of intermediate toxicity and kinetic efficiency in the regulation of prokaryotic metabolism
PLOS Computational Biology 13, 2017, e100537 - S. Dühring, S. Germerodt, C. Skerka, P. F. Zipfel, T. Dandekar, S. Schuster
Host-pathogen interactions between the human innate immune system and Candida albicans - Understanding and modeling defense and evasion strategies
Frontiers in Microbiology 6 (625), 2015
- J. Ewald, P. Sieber, R. Garde, S.N. Lang, S. Schuster, B. Ibrahim
-
Funding
DFG Collaborative Research Center / Transregio 124 "FungiNet"
-
Project description
Fast growing cells like cancer cells, show the phenomenon of an incomplete metabolization of glucose to lactate, called Warburg effect in cancer cells. Despite its lower energy yield per molecule compared to the full respiration producing CO2, linear programming models show that this metabolic route is optimal with regard to enzyme costs and energy rate. In extended models we are investigating additional energy sources like glutamine to understand the uptake of other carbon sources by cancer cells.
-
Literature
- J. Ewald, Z. He, W. Dimitriew, S. Schuster
Including glutamine in a resource allocation model of energy metabolism in cancer and yeast cells.
npj Systems Biology and Applications 10, 77 (2024). - P. Möller, X. Liu, S. Schuster, D. Boley
Linear programming model can explain respiration of fermentation products.
PloS one 13, 2018, e0191803 - S. Schuster, D. Boley, P. Möller, H. Stark, C. Kaleta
Mathematical models for explaining the Warburg effect: A review focussed on ATP and biomass production.
Biochemical Society Transactions 43 (6), 2015, 1187-1194 - S. Schuster, L. de Figueiredo, A. Schroeter, C. Kaleta
Combining Metabolic Pathway Analysis with Evolutionary Game Theory. Explaining the occurrence of low-yield pathways by an analytic optimization approach.
BioSystems 105, 2011, 147-153
- J. Ewald, Z. He, W. Dimitriew, S. Schuster
-
Project description
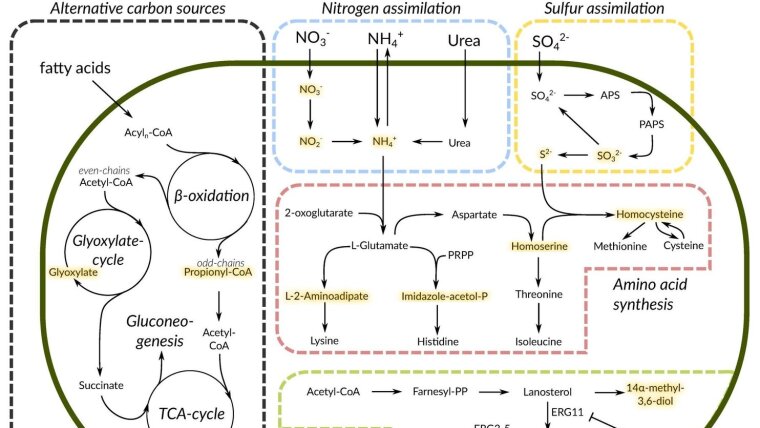
Genome-scale metabolic models are useful in that they can provide insight into how the multitude of reactions in living cells is interconnected, and how changes in some reactions can affect the entire network. We are focused on the construction of a genome-scale metabolic model of a pathogenic fungus Lichtheimia corymbifera. According to WHO, this fungus belongs to a group of human pathogens with a high priority, and is capable of causing severe diseases. At the same time, the fungus is actively used as a fermentation agend in many cultures, without obvious side effects. Our ultimate goal is to explain this discrepancy based on the available data regarding the metabolism of L. corymbifera.
In this work, we use toolboxes for genome-scale model reconstructions, metabolism-related databases and packages for interaction with them, as well as large language models to comprehend thousands of reactions that occur in the living organism.
-
Literature
To be added