-
Schuster, Stefan, Univ.-Prof. Dr Professorship of Bioinformatics
Room 3013
Inselplatz 5
07743 Jena -
Wesp, Valentin Professorship of Bioinformatics
Room 2008
Inselplatz 5
07743 Jena
Image: Fotostudio Gebhardt Jena -
Malycheva, Tatiana Professorship of Bioinformatics
Room 2007
Inselplatz 5
07743 Jena
Image: Fotostudio Müller -
Lovate, Gabriel Lencioni Professorship of Bioinformatics
Room 2008
Inselplatz 5
07743 Jena
Image: Alfonso (Poncho) Aceves-Aparicio
Protein aging
-
Project description
The focus of the project lies on age-related protein damage that leads to aggregates. These cannot be degraded anymore and therefore accumulate until the cell suffers from major functional inhibitions and finally triggers apoptosis. The goal is to determine what are the most or also least susceptible targets in terms of oxydative damage. Also information from structure and prominent aggregate proteins will be included. Furthermore functional connections and domains will be examined in this context.
-
Literature
- M. Fichtner, S. Schuster, H. Stark
Influence of spatial structure on protein damage susceptibility: a bioinformatics approach
Sci. Rep. 11, 2021, 4938 - E. Barth, P. Sieber, H. Stark, S. Schuster
Robustness during Aging — Molecular Biological and Physiological Aspects
Cells 9 (8), 2020, 1862 - M. Fichtner, S. Schuster, H. Stark
Determination of scoring functions for protein damage susceptibility
BioSystems 187, 2020, 104035, epub ahead of print - S. Schäuble, K. Klement, S. Marthandan, S. Münch, I. Heiland, S. Schuster, P. Hemmerich, S. Diekmann
Quantitative model of cell cycle arrest and cellular senescence in primary human fibroblasts.
PLoS ONE 7, 2012, e42150
- M. Fichtner, S. Schuster, H. Stark
Co-Evolution of Antibiotic Resistance and Biosynthetic Gene Clusters
-
Project description
Antibiotic resistance genes (ARGs) and biosynthetic gene clusters (BGCs) play crucial roles in microbial communities by influencing antimicrobial production and adaptation to selective pressures. In this project, we investigate how environmental disturbance, microbial competition, and genetic mobility drive ARG-BGC coevolution across ecosystems. We will analyze metagenomic datasets from pristine and anthropogenically impacted sites using a custom Nextflow pipeline that integrates BGC detection tools (antiSMASH) with ARG detection frameworks (AMRFinderPlus, MEGARes, ResFinder). After validation, we'll expand to other datasets to assess genomic trait dissemination under different environmental conditions. Initially focused on metagenomics, we plan to eventually incorporate metatranscriptomics and metabolomics approaches. Our findings could improve antibiotic stewardship strategies and highlight the value of pristine habitats as reservoirs for next-generation antimicrobials.
-
Literature
- T.C. Borelli, G.L. Lovate, A.F.T. Scaranello, L.F. Ribeiro, L. Zaramela,F. M. Pereira-dos-Santos, R. Silva-Rocha, M.E. Guazzaroni
Combining Functional Genomics and Whole-Genome Sequencing to Detect Antibiotic Resistance Genes in Bacterial Strains Co-Occurring Simultaneously in a Brazilian HospitalExternal link
Antibiotics 10 (4), 419 - L.F. Alves, C.A. Westmann, G.L. Lovate, G.M.V. de Siqueira, T.C. Borelli, M.E. Guazzaroni
Metagenomic approaches for understanding new concepts in microbial scienceExternal link
International journal of genomics 2018 (1), 2312987
- T.C. Borelli, G.L. Lovate, A.F.T. Scaranello, L.F. Ribeiro, L. Zaramela,F. M. Pereira-dos-Santos, R. Silva-Rocha, M.E. Guazzaroni
Classification and combinatorics of amino acids and fatty acids.
-
Project description
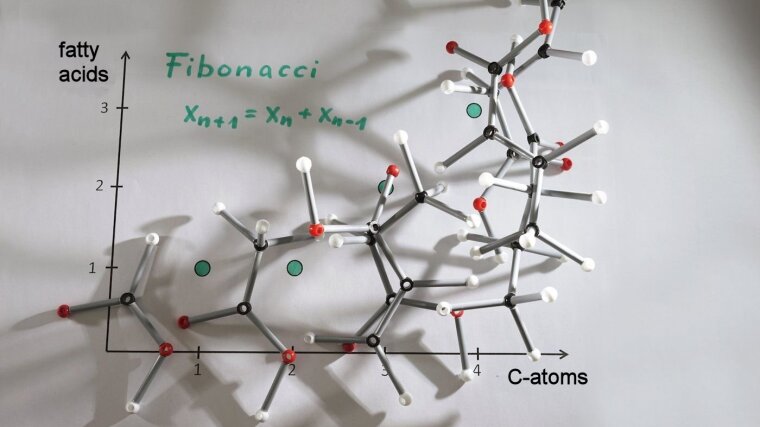
The first part of this project consists of investigating the combinatorial possibilities of aliphatic amino acids and fatty acids. For this purpose molecular classes with certain constrictions are chosen because an exhausting listing is not feasible. Mathematical computations as for example recursions on the resulting number sequences are conducted. One possible question is how many unbranched, unmodificated fatty acids with n C-atoms principally can exist. This leads to the fibonacci numbers. Additionally literature research is conducted to verify which combinations are actually synthesized by real organisms. The second part of the project deals with the development of novel classification attempts for amino acids to improve or supplement existing approaches. As we hope, this leads to novel insights into key questions of biochemical evolution.
-
Literature
- S. Schuster, T. Malycheva
Enumeration of saturated and unsaturated substituted N-heterocycles
arXiv:2309.02343, 2023, Preprint - A. Then, H. Zhang, B. Ibrahim, S. Schuster
Bioinformatics analysis of the periodicity in proteins with coiled-coil structure—Enumerating all decompositions of sequence periods
Intern. J. Molec. Sci. 23, 2022, 8692 - A. Then, K. Mácha, B. Ibrahim, S. Schuster
A novel method for achieving an optimal classification of the proteinogenic amino acids
Scientific Reports 10, 2020, 15321
https://www.nature.com/articles/s41598-020-72174-5External link - M. Fichtner, K. Voigt, S. Schuster
The tip and hidden part of the iceberg: Proteinogenic and non-proteinogenic aliphatic amino acids
Biochimica and Biophysica Acta - General Subjects 1861 (1, Part A), 2017, 3258-3269
https://www.ncbi.nlm.nih.gov/pubmed/27554846External link - S. Schuster, M. Fichtner, S. Sasso
Use of Fibonacci numbers in lipidomics - Enumerating various classes of fatty acids
Scientific Reports 7, 2017, 39821
https://www.ncbi.nlm.nih.gov/pubmed/28071669External link - K. Grützmann, S. Böcker, S. Schuster
Combinatorics of aliphatic amino acids
Naturwissenschaften 98, 2011, 79-86
https://link.springer.com/article/10.1007/s00114-010-0743-2External link
- S. Schuster, T. Malycheva
Alternative splicing in fungi
-
Project description
As in higher eukaryotes, fungal species can splice their mRNA in an alternative (differential) manner. But how wide-spread is this cellular process in the fungal kingdom and which processes of the microbial lifestyle are affected? Does alternative splicing facilitate multicellularity? Does it affect the switch from peaceful mutualism to pathogenic behavior?
To address these issues, we analyze RNA-Seq data of various fungi, such as Candida albicans, Aspergillus fumigatus, and Cryptococcus neoformans. Several analysis tools are applied in order to identify (unknown) alternative splicing events. Detected alternatively spliced genes are viewed in a cell biological context. We study conservation of these genes and investigate phylogenetic distribution. -
Literature
- P. Sieber, M. Platzer, S. Schuster
The definition of open reading frame revisited
Trends in Genetics 34, 2018, 167-170 - V. Wesp, G. Theißen, S. Schuster
Statistical analysis of synonymous and stop codons in pseudo-random and real sequences as a function of GC content
Sci. Rep. 13, December, 2023, 22996 - P. Sieber, E. Barth, M. Marz
The landscape of the alternatively spliced transcriptome remains stable during aging across different species and tissues
bioRxiv February, 2019, 541417 - P. Sieber, K. Voigt, P. Kämmer, S. Brunke, S. Schuster, J. Linde
Comparative study on alternative splicing in human fungal pathogens suggests its involvement during host invasion
Frontiers in Microbiology 9, 2018, 2313 - P. Sieber, M. Platzer, S. Schuster
The definition of open reading frame revisited.
Trends in Genetics 34, 2018, 167-170 - J. Linde, S. Duggan, M. Weber, F. Horn, P. Sieber, D. Hellwig, K. Riege, M. Marz, R. Martin, R. Guthke, O. Kurzai
Defining the transcriptomic landscape of Candida glabrata by RNA-Seq.
Nucleic acids research 43, 2015, 1392-1406
- P. Sieber, M. Platzer, S. Schuster