Genome-wide approaches like high-throughput DNA- and RNA-sequencing have shown that only a small part of eukaryotic genomes codes for proteins (e.g. in human only 1.5%). However, virtually the whole non-coding part is also transcribed, giving rise to thousands of non-coding RNAs (ncRNAs). The biological role of these ncRNAs has been intensively investigated over last decade, showing that they have various functions in the cell nucleus and the cytoplasm. One of the most interesting features of ncRNAs is that many of them can modulate the chromatin state in a site-specific manner. However, in most cases it is not clear how ncRNAs find their specific target sites in the genome.
Tethering via chromatin-bound proteins can be one mechanism, but is indirect and has limited discriminative power. Therefore, direct sequence-specific interactions of ncRNAs with genomic DNA are better suited. In fact, nascent RNA can displace the non-template DNA strand and base-pair with the template strand, leading to a RNA/DNA hybrid and a single stranded DNA, a structure which is termed R-loop. R-loops have been shown to have indeed regulatory functions, but they depend on ongoing transcription and expose the single strand to an increased risk of DNA damage.
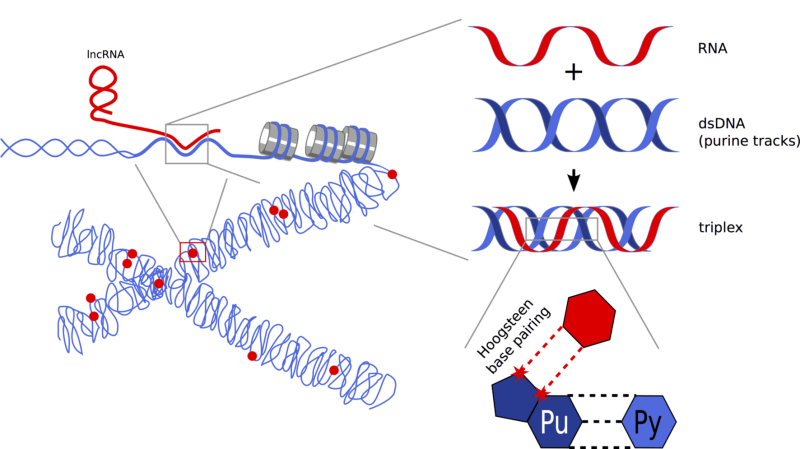
Another triple stranded RNA/DNA structure are triple helices (triplexes), in which the RNA binds to the major groove of the DNA double helix via Hoogsteen base pairing. RNA:DNA triplexes have certain sequence requirements and thus allow a relaxed sequence specificity between both nucleic acids, i.e. triplex-forming RNAs can act in cis or in trans at one or several genomic loci. These features, together with the non-invasive interaction with DNA, make RNA:DNA triplex formation a very attractive targeting mechanism for ncRNAs. However, the detection of RNA:DNA triplexes in cells is very difficult and there are only a few reports describing a role of these structures in gene regulation.
One major aim of our work is to comprehensively identify and characterize ncRNA:DNA triplexes. We develop tools and techniques to detect these structures in cells that will allow us to globally map triplex-forming sites in the genome. From these data we hope to deduce general rules of triplex-formation. To decipher the role of ncRNA:DNA triplexes in shaping the chromatin landscape, we study individual triplexes under different physiological conditions and manipulate triplex formation by knocking down (loss-of-function) or overexpressing (gain-of-function) the respective ncRNAs. Moreover, we investigate how triplexes can compete with binding of proteins or with formation of alternative nucleic acid structures at the target sites, but also how (and which) chromatin-associated factors are recruited by triplexes. Together, we hope that our work will provide a profound understanding of ncRNA-mediated triplex-formation, which, according to the vast number of genomic sites with triplex-forming potential, is an important and abundant mechanism of epigenetic regulation.
Triplex
Image: Holger Bierhoff