Introduction
Susie Grigson at Flinders University
Image: Susie GrigsonI am a visiting PhD student from Flinders University in Australia.
Approximately 30% of bacterial and 65% of viral protein sequences cannot be assigned a known biological function. To gain a better understanding of these microbes, my PhD research aims to develop new computational techniques and tools to decrease this ever-increasing sequence function gap. I use machine learning, sequence embeddings and computed protein structures combined with a range of genomic properties including gene arrangements to build software which allows researchers to annotate unknown microbial genes.
Career path
- Flinders University, South Australia; Bachelor of Science (Molecular Biology and Biochemistry) 2018-2020
- Flinders University, South Australia; Bachelor of Science (Mathematics)(Honours) 2017-2021
Thesis: Dynamics of Microbial Communities During Continual Migration - Flinders University, South Australia; Doctor of Philosophy (Bioinformatics) 2022-2025
Thesis: Computational methods for predicting microbial protein functions - Friedrich Schiller University Jena; Visiting Research Student 2023
Representative research
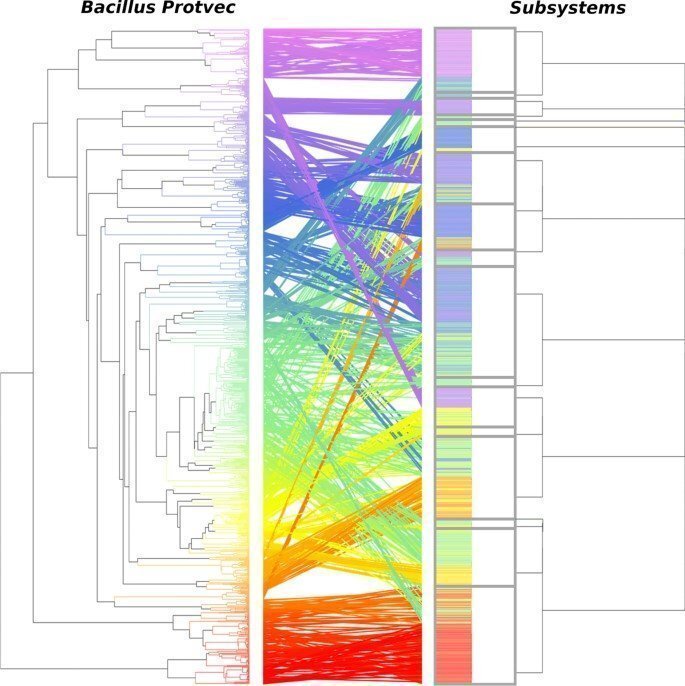
I have investigated how amino acid sequence embeddings may be used to study functional hierarchies for describing bacterial protein functions. Using a Protvec sequence embedding, a technique used to represent amino acid sequences in n-dimensional space, I demonstrated that the bacterial carbohydrate metabolism class within the SEED annotation system contains 48 clusters of embedded sequences. However, these sequences are currently described using 29 functional labels arranged within a hierarchy which is different to the hierarchical organisation of sequences within the Protvec embedding. Furthermore, by representing unknown sequences with Protvec, I demonstrated that unknown sequences form clusters that likely share related biological roles. Such clusters may be beneficial for selecting optimal candidate proteins to characterize experimentally.
Doi: https://doi.org/10.1186/s12859-022-04930-5External link
Comparison of Bacillus carbohydrate metabolism sequences grouped using agglomerative clustering on sequence embeddings using the Bacillus carbohydrate metabolism Protvec model and the SEED annotation hierarchy. The color joining the dendrograms is continuous across the Protvec dendrogram. Boxes are drawn around each subsystem in the SEED annotation hierarchy.
Illustration: Susie GrigsonPublications
- Susanna R. Grigson, Jody C. McKerral, James G. Mitchell, and Robert A. Edwards (2022): "Organizing the bacterial annotation space with amino acid sequence embeddings"External link, BMC bioinformatics 23(1): 385, doi: https://doi.org/10.1186/s12859-022-04930-5External link.
- Suzanne Scott, Susanna Grigson, Felix Hartkopf, Claus V. Hallwirth, Ian E. Alexander, Denis C. Bauer, and Laurence OW Wilson (2022): "A bioinformatic pipeline for simulating viral integration data."External link Data in Brief 42: 108161, doi: https://doi.org/10.1016/j.dib.2022.108161External link.
- Suzanne Scott, Claus V. Hallwirth, Felix Hartkopf, Susanna Grigson, Yatish Jain, Ian E. Alexander, Denis C. Bauer, and Laurence OW Wilson (2022): "Isling: a tool for detecting integration of wild-type viruses and clinical vectors."External link Journal of molecular biology 434(11):167408, doi:https://doi.org/10.1016/j.jmb.2021.167408External link.
- Bhavya Papudeshi, Alejandro A. Vega, Cole Souza, Sarah K. Giles, Vijini Mallawaarachchi, Michael J. Roach, Michelle An Nicole Jacobson, Katelyn McNair, Maria Fernanda Mora, Karina Pastrana, Lance Boling, Christopher Leigh, Clarice Harker, Will S. Plewa, Susanna R. Grigson, George Bouras, Przemysław Decewicz, Antoni Luque, Lindsay Droit, Scott A. Handley, David Wang, Anca M. Segall, Elizabeth A. Dinsdale and Robert A. Edwards (2023): "Host interactions of novel Crassvirales species belonging to multiple families infecting bacterial host, Bacteroides cellulosilyticus WH2."External link bioRxiv 2023-03, doi: https://doi.org/10.1101/2023.03.05.531146External link.
- Vijini Mallawaarachchi, Michael J. Roach, Bhavya Papudeshi, Sarah K. Giles, Susanna R. Grigson, Przemyslaw Decewicz, George Bouras, Ryan D. Hesse, Laura K. Inglis, Abbey L. K. Hutton, A. Dinsdale and Robert A. Edwards (2023): "Phables: from fragmented assemblies to high-quality bacteriophage genomes."External link bioRxiv: 2023-04, doi: https://doi.org/10.1101/2023.04.04.535632External link.