- Light
- Life
- Liberty
- Publications
- Research
- Team
Published: | By: Piotr Rozwalak and Marcel Baecker
Can we reconstruct bacteriophage genomes from millennia-old archaeological samples like paleofeces? This question has driven the research of our PhD candidate, Piotr Rozwalak, for the past two years. Today, we proudly announce his pioneering publication on ancient viruses in Nature CommunicationsExternal link. We asked Piotr to share his personal story behind this publication below.
In autumn 2021, as a geology and biology graduate, I sought a topic for my bioinformatics master's thesis with Andrzej ZielezińskiExternal link, one of the best bioinformaticians at Adam Mickiewicz UniversityExternal link in Poznań, Poland. He develops computational tools to predict hosts for viruses. However, my previous interest focused on ancient DNA, especially from sediments, and I had no experience with virology. Despite this, I ventured into a research project about detecting bacteriophage sequences in ancient metagenomes.
Most ancient metagenomics focused on studying bacterial genomes and ignored the viral sequences present in those same datasets. Despite challenges with degraded DNA, we produced promising results in recovering ancient bacteriophage genomes, which I followed up on during my first visit to Jena for an Erasmus traineeship program.
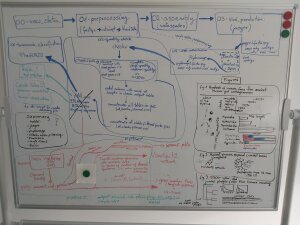
Whiteboard with project plan
Image: PiotrThe VEO Group goes Paleo!
As a young geologist, I was used to spending long periods in the field collecting samples for analysis. However, the summer of 2022 was radically different. Instead of being outdoors, I found myself in front of my computer, mining genomes from paleofeces and gut contents of people who lived thousands of years ago. During my 2-month visit to Jena, I worked on developing a workflow (see image) to study ancient phages in the newly founded VEO Group. At the end of my visit, with the help of my colleagues, I already had a collection of authenticated ancient phage genomes. Among this viral genomic diversity, I was amazed to find a single, complete genome sequence that was 97.7% identical to its closest modern reference. It contradicted the standard view that phages evolve quickly, driven by competition with their bacterial hosts. In the months following my traineeship in Jena, we described this phenomenon of evolutionary stasis in detail, but there were no final answers yet.
We published a preprint of our manuscript in June 2023 on bioRxiv, and the day after, we received an email from the Editor of Nature Communications with an invitation to submit our paper to this journal. Naturally, I was thrilled, and after a few days, we responded to the invitation and submitted our article. The reviewers were kind and helpful, and after some improvements, the article was finally accepted and published in January 2024. We are delighted Senior Editor César Sánchez selected our paper as an Editor’s highlightExternal link in the Microbiology and Infectious Diseases section of Nature Communications.
I was surprised and happy to find myself in popular demand for interviews about my work. Both of my universities, Jena and PoznańExternal link, and a countrywide portalExternal link popularizing science in Poland interviewed me for their websites. Local news agencies like Jena TVExternal link or radio PoznańExternal link wrote articles about me and my paper. The Polish Ministry of Science has honored me with a grant for young researchers, enabling me to explore ancient bacteriophages further. In Jena, we were lucky to discuss our research with colleagues from the Microverse clusterExternal link, including renowned Archeogeneticists Prof. Christina Warriner (MPI, HKI, FSU, HarvardExternal link) and Paleobiotechnologist Prof. Pierre Stallforth (HKIExternal link), to identify future research avenues of ancient viromics. I look forward to continuing this line of research and making an impact on paleoviromics.
Original Publication
Rozwalak, P., Barylski, J., Wijesekara, Y. et al. Ultraconserved bacteriophage genome sequence identified in 1300-year-old human palaeofaeces. Nat Commun 15, 495 (2024). https://doi.org/10.1038/s41467-023-44370-0External link